Project Organization and Data Lifecycle
When setting up a project on the Tufts HPC it is worth considering what kind of resources you need and the data’s lifecyle:
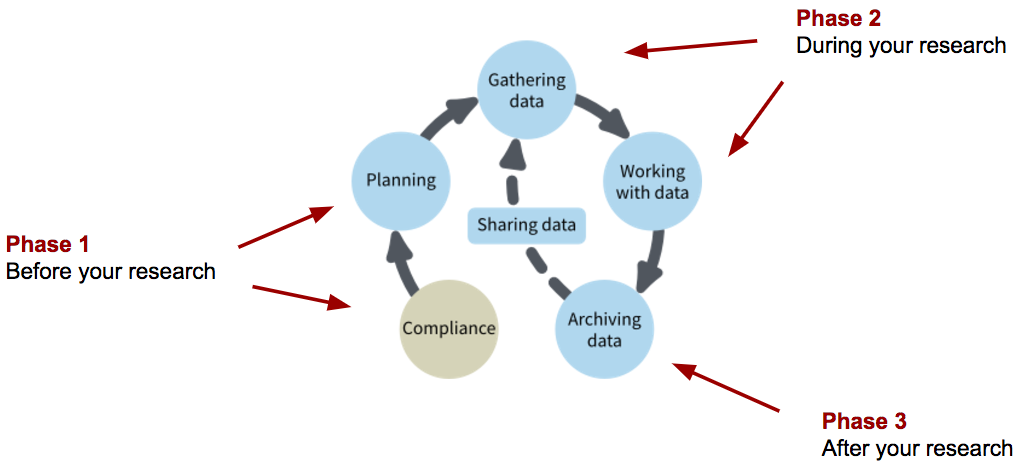
Questions To Ask
When considering compute resources make sure you have an idea of the following:
| Question | Why it’s Useful |
|---|---|
| What is the approximate start date and end date for usage of the data? | This can be helpful when it comes to how TTS plans future storage needs |
| What is the estimated computation needed? | Knowing how long your jobs might take is incredibly valuable for planning when you’ll run them. If you know the job takes 2 days to run, you’ll be able to plan accordingly. |
| Are you working with multiple files simultaneously? | Each directory comes with some limit. You can ask for more storage but often storage is stretched thin as it is. If you can stagger when large files are loaded you can avoid hitting your storage maximum. |
| Can you remove redundant files? | Life sciences data is rife with redundant files/logs. Consider if there is anything that can be deleted or even downloaded again if it is a publically available file. |
| What software/tools will you use to process, analyze, or view the data? | The Tufts HPC clustser has an incredible array of tools to help you do research. However, it doesn’t have all of them and it might help to check before you start your project. |
Project Organization
It might seem insignificant but consider creating an organized folder structure. As a project grows it can be difficult to find relavent data even when the folder structure is neat. Consider the following two projects and how easy it might be to find the files you’re looking for:
Project 1
[tutln01@c1cmp047 project1]$ ls
15338084.err 15338085.err 15338086.err 15338087.err data sraAccList.txt SRR19145573.fastq.gz STAR1 starLoop.sh
15338084.out 15338085.out 15338086.out 15338087.out download.sh SRR19145569.fastq.gz STAR starAll.sh starOne.sh
Project 2
[tutln01@c1cmp047 project1]$ ls
input_data trimmed_data bam_files feature_counts DEG_results