Description
This course is an introduction to RNA sequencing using Galaxy on Tufts University’s Galaxy server. For support with this workshop or Galaxy, see our Galaxy documentation or email TTS Research Technology tts-research@tufts.edu
Goals
Bioinformatics for RNAseq has the following workflow steps
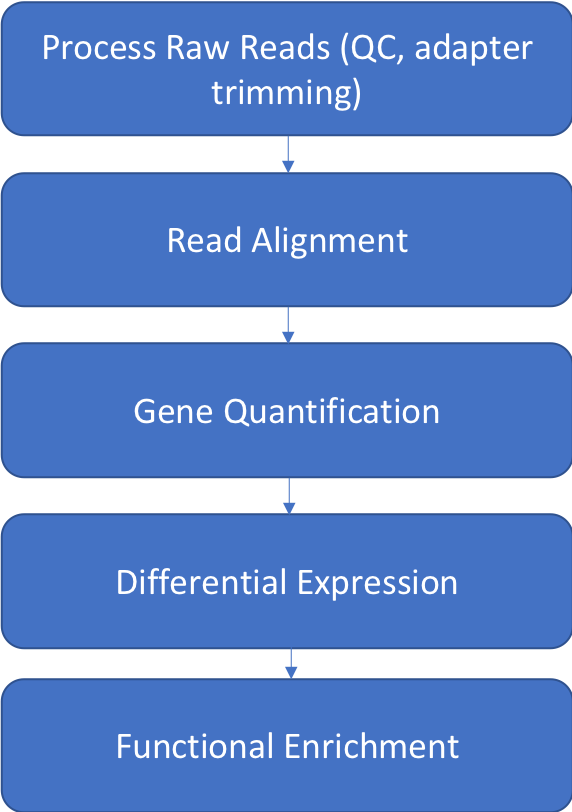
Materials Needed
- Chrome web browser
- Account on Tufts High Performance Compute Cluster
- VPN if accessing the HPC from off campus
Table of Contents
- Introduction Slides
- Introduction to Galaxy
- Next: Process Raw Reads
- Read Alignment
- Gene quantification
- Differential Expression
Acknowledgement
Thanks to bioinformatics intern Meg Farely for help developing this workshop. Much of this explanation has been adapted from these sources:
- Harvard Chan Bioinformatics Core RNAseq training
- DESeq2 Vignette
- Introduction To RNA-seq with Galaxy


