Intro to RNA Seq
Description:
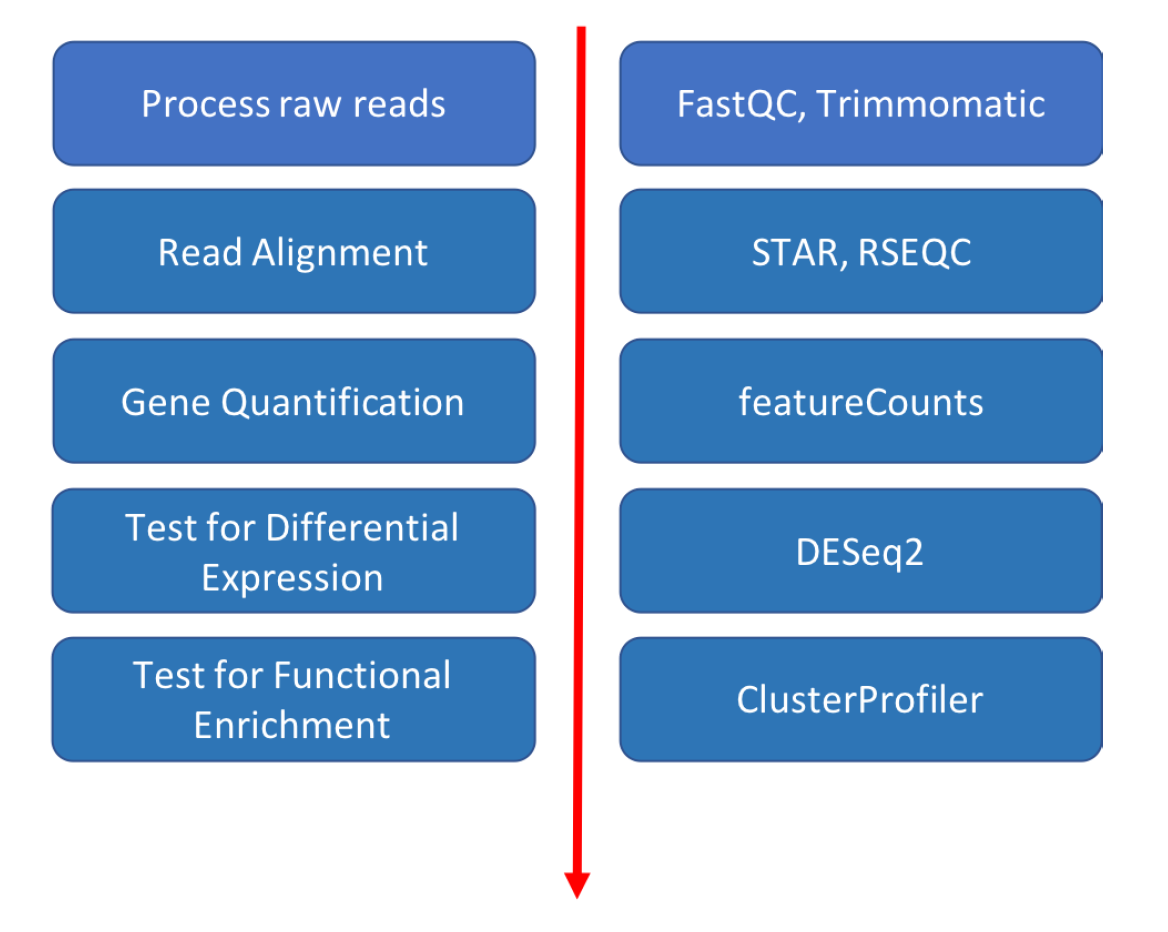
This tutorial will cover the basics of bioinformatics for RNA sequencing analysis using command line tools and R on the Tufts High Performance Compute Cluster (HPC). The analysis pipeline includes quality control, read alignment, feature quantification, differential expression and pathway analysis. There was a 1-hour introductory session, after which students will follow this self-guided online material. The material is designed to run on Tufts High Performance Compute (HPC) Cluster. If you don’t have access to Tufts HPC, you will need to install all required modules on your own computer.
Goals
- Intro to RNA sequencing logistics
- Write and run bash scripts
- Intro to command line bioinformatics tools: FastQC, STAR, SAMtools
- Perform differential expression analysis using DESeq2 in Rstudio
Prerequisites
Basic understanding of computational skills
- Introduction to Linux
- Introduction to HPC
- Introduction to R workshop materials and recording
Materials Needed
If you are a Tufts member and have access to Tufts HPC:
If you don’t have access to Tufts HPC:
All scripts you will be using:
1-hr introduction session slides:
Schedule
- Currently at: Course Home
- Introduction
- Setup using Tufts HPC
- Quality Control
- Read Alignment
- Gene Quantification
- Differential Expression
- Pathway Enrichment
Acknowledgement
The original workshop repository can be found here. Much of this workshop was adapted from Bioinformatics @ Tufts and the HBC Training DGE workshop with the help of Dr. Rebecca Batorsky and Dr. Albert Tai at Tufts University.