Intro to Variant Calling
This course covers the basics of Variant Calling and interpretation for Next Gen sequencing data.
The material is designed to be a 3 hour workshop and is run on Tufts High Performance Compute (HPC) Cluster. For support with this workshop or the HPC cluster, email TTS Research Technology tts-research@tufts.edu
If you are taking this workshop on 5/19/20, please use Piazza for discussion. The class is listed in term Summer 2020, 1: Intro to NGS Bioinformatics
Goals
- Writing and running bash scripts on the HPC
- Intro to several bioinformatics tools: BWA, Samtools, Picard, GATK, VEP, IGV
- Variant Calling, Annotation and Interpretation using a Human Exome sample
Prerequisites
Computational skills needed
Materials Needed
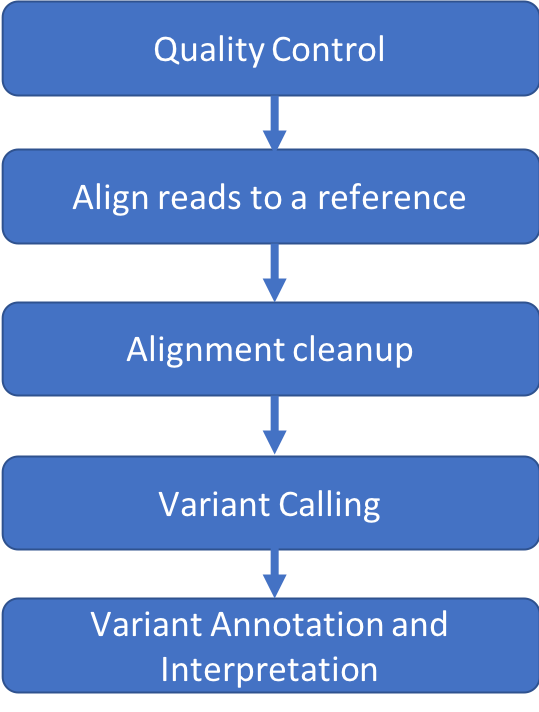
Schedule
- Introduction
- Setup
- Quality Control
- Alignment
- Alignment Cleanup
- Variant Calling
- Annotation and Interpretation
Acknowledgement
This course was developed by Dr. Wewen Huo and Dr. Rebecca Batorsky at Tufts University and adapted from HBC Training In Depth NGS Analysis Course
The original course repo can be found here


